Analysis of Calcium Imaging Data
Training materials for V4SDB Student Winter School
Instructor:
Isaac Bianco (University College London)
i.bianco@ucl.ac.uk
Workshop Overview
In vivo imaging of calcium dynamics can shed light on a broad range of biological processes from cell migration and tissue morphogenesis to multiple aspects of neuronal activity. In neuroscience, a combination of modern genetically encoded calcium sensors alongside advances in imaging technology are enabling the activity of tens of thousands of neurons to be monitored in behaving animals. In parallel, advances in data processing and analysis are enabling researchers to make sense of vast population activity datasets.
In this workshop, we will:
- introduce the key principles of calcium imaging;
- learn how to process raw calcium imaging data and extract estimates of neuronal activity;
- examine ways to explore large population activity datasets and relate neural dynamics to sensory stimuli, task variables, and animal behaviour.
Workshop slides will be available here
Software
Fiji:
- An open-source image processing package.
- follow the instructions under Local Installation
- be sure to install the GUI version:
python -m pip install suite2p[gui] - on the Mac you may need to use:
python -m pip install 'suite2p[gui]'
- follow the instructions under Local Installation
- again, be sure to install the GUI version
- it should be fine to install it in the same suite2p environment as used above
Datasets
a small calcium recording that we will look at in FIJI link
(You will need to click on theDownload raw filebutton to download the dataset.)a large calcium imaging movie for Suite2p testing
- download any one of the TIFF stacks in this folder link
a zebrafish calcium imaging movie for Suite2p testing link
a zebrafish activity dataset for Rastermap testing link
- plus some extra files to help interpret the fish dataset link
Install the Python environment
- Install an Anaconda distribution of Python
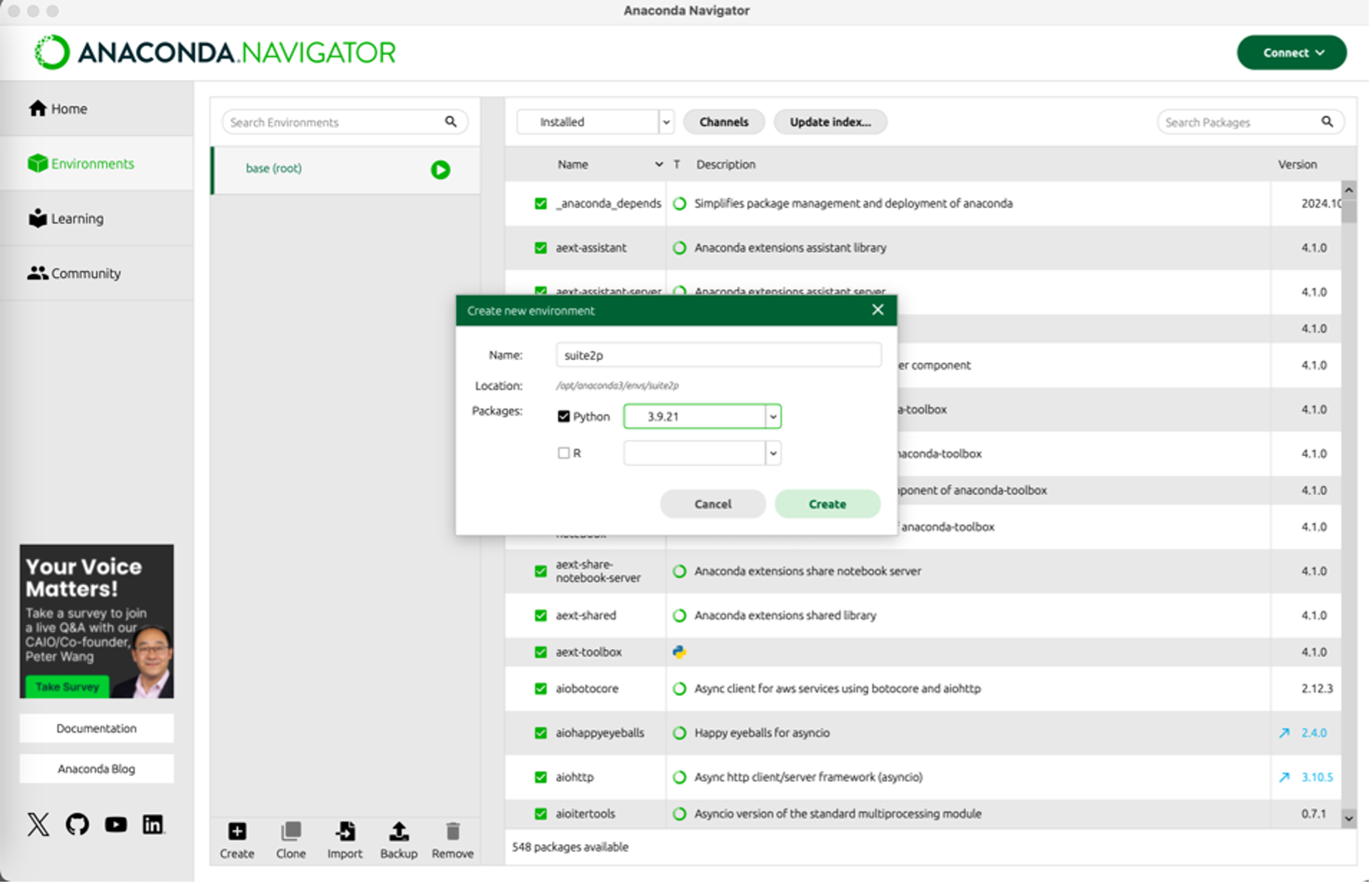
- Using either the command line or the Anaconda Navigator, create a new environment
- Command line:
conda create --name suite2p python=3.9or - Anaconda Navigator, see pic below
- Command line:
- Using the command line (open “Terminal” on the Mac), activate the suite2p environment:
conda activate suite2p
Now install suite2p by typing:
python -m pip install suite2p[gui]or on the Mac you might need:
python -m pip install ‘suite2p[gui]’(notice the ‘ )

- Once the download is finished, launch suite2p:
python -m suite2p

All done!
References
Suite2p
Pachitariu M, Stringer C, Dipoppa M, et al. (2017) Suite2p: beyond 10,000 neurons with standard two-photon microscopy. bioRxiv https://doi.org/10.1101/061507
Suite2p documentation: https://suite2p.readthedocs.io/en/latest/
OASIS
Friedrich, J., Zhou, P., & Paninski, L. (2017). Fast online deconvolution of calcium imaging data. PLOS Comp Bio, 13(3), e1005423. https://doi.org/10.1371/journal.pcbi.1005423
Rastermap
Stringer, C., Zhong, L., Syeda, A., Du, F., Kesa, M., & Pachitariu, M. (2025). Rastermap: a discovery method for neural population recordings. Nat Neurosci, 28(1), 201–212. https://doi.org/10.1038/s41593-024-01783-4
